|
0 Comments
While reviewing SARS-CoV-2 literature, serendipitously came across a new MexRXiv posting by the Arnaout laboratory titled: "Contrasting Effects of SARS-CoV-2 Vaccination vs. Infection on Antibody and TCR Repertoires." Fascinating contribution to the understanding our immune response to this infection. Congratulations, Dr. Arnaout!
On the "Susceptibility and resistance of multidrug-resistant gram-negative bacteria to novel beta-lactam/beta-lactamase inhibitor combinations"!!! Way to go Dr. Brennan-Krohn
Just out our recently posted pre-print in MedRXiv --> real-world data to inform regulatory pathways for SARS-CoV-2 PCR and antigen tests from our work led by Clin Micro Attending, Ramy Arnaout, sponsored by the Reagan-Udall foundation.
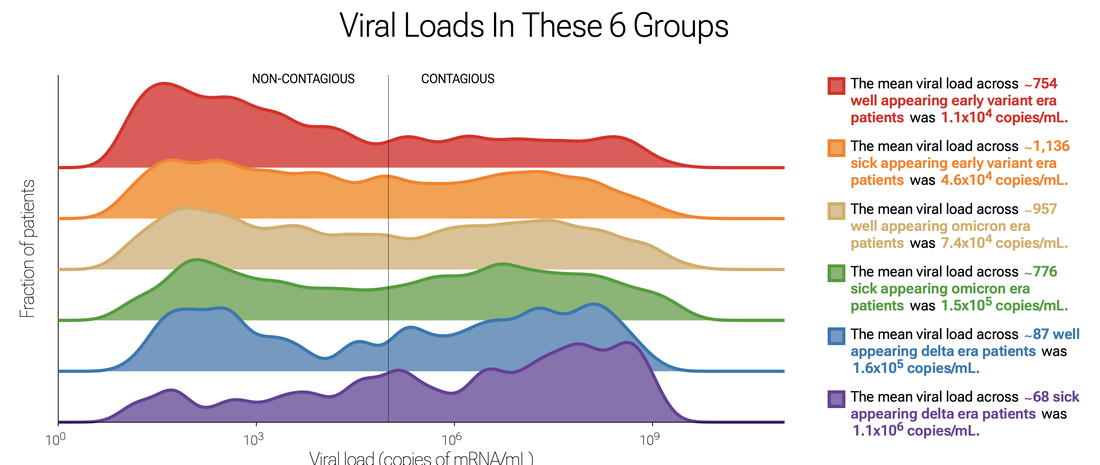
Check out the data yourself - how any of large number of metadata influences SARS-CoV-2 testing results. An example below is a plot of viral loads by variant and whether patients were sick or well. Delighted to learn that Thao Truong, PhD, who performed a three month rotation in our clinical microbiology laboratory and participated in our translational research efforts, later obtaining a CPEP fellowship position at UCLA Children's Hospital, is now a clinical microbiology faculty member at the University of Washington! Way to go, Thao!
See our study in collaboration with the Kanki Laboratory at the Harvard School of Public Health. We used live cultured WA1, Omicron and Delta to compare limits of detection. Live virus was used to avoid artifact potentially introduced by use of gammma-irradiated or heat-inactivated virus used by manufacturer's in their submission to the FDA for EUA designation. Omicron detected just as well as WA1. Delta less well. Published in Journal of Clinical Microbiology today
Congratulations to Clin Micro Faculty Member, Ramy Arnaout: https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0265233journals.plos.org/plosone/article?id=10.1371/journal.pone.0265233
This $900,000 award is to examine association of real-world data with SARS-CoV-2 PCR and antigen test performance and advise the FDA on streamlining its regulatory approach for these tests. Submitting PI clin micro faculty Ramy Arnaout. Co-PI's, Phyllis Kanki (HSPH), and clin micro faculty Stefan Riedel and James Kirby. Announcement: Reagan-Udall Foundation for the FDA Awards $1.8 Million in Research Funding for Studies on the Real-World Performance of COVID-19 Tests
See our Preprint just posted. Not consistent with statement on FDA website indicating that Omicron detected less efficiently.
Check out the visualization of the relationship of mortality versus case numbers during different ways of the pandemic by country:
|
 RSS Feed
RSS Feed